🧬 ChemMiniQ3 - with Horizon Loss on SELFIES and Biologically-Aware RL Fine-Tuning
A lightweight experimental generative model for chemistry, built on mini Qwen2-like with multi-horizon predictive loss for molecular SELFIES representations.
Disclaimer: For Academic Purposes Only
The information and model provided is for academic purposes only. It is intended for educational and research use, and should not be used for any commercial or legal purposes. The author do not guarantee the accuracy, completeness, or reliability of the information. Prototype research code — not production-ready. Learning by building.
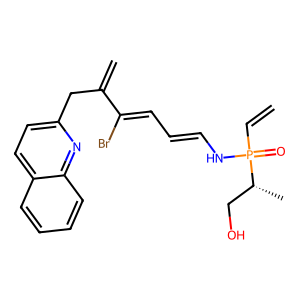
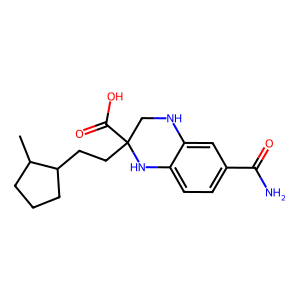
A custom Qwen2-style language model, adapted for molecular generation:
- ✅ Qwen2-like Architecture – Modernized backbone with efficient attention
- ✅ Multi-Token Prediction (MTP Head) – Predicts multiple future tokens (1–3) in parallel
- ✅ Horizon Loss – Weighted multi-horizon objectives for longer-term sequence coherence
- ✅ SELFIES-native Tokenizer – Robust encoding for valid molecular structures with FastChemTokenizer
- ✅ Ranger21 Optimizer – Adaptive optimizer with warmup/warmdown scheduling
- ✅ Gradient Checkpointing – Trainable on smaller GPUs
- ✅ Streaming Dataset Loader – Trainable on smaller RAM
Experimental RL PPO-KL-ready features:
- ✅ Enhanced Reward Functions – Validity, Lipinski, charge neutrality, diversity, complexity
- ✅ Curriculum Learning – Gradually increases generation length during training
- ✅ Adaptive KL + Entropy Control – Stabilizes reinforcement learning fine-tuning
💡 Target domain: chemistry & molecular generation (SELFIES).
🚀 Architecture is potentially generalizable to other sequence domains.
Pre-trained model's (with-RL) description:
Model has 9,854,851 trainable parameters.
Input shape: torch.Size([2, 32])
Logits shape: torch.Size([2, 32, 782])
Trained on 14k samples from combined curated dataset built from COCONUTDB (Sorokina et al., 2021),
ChemBL34 (Zdrazil et al., 2023), and SuperNatural3 (Gallo et al. 2023) dataset
Batch Size: 16 (* 4 Grad acc -> ~64)
Optimizer: Ranger21 (MADGRAD-Lookahead-AdaBelief with gradient centralization, linear warm up (22%),
gradient clipping, and L2 weight decay)
Learning rate: 5e-06 (**Warmup complete - lr set to 3.9e-06)
Training log for E-1:
Warm-up
time step loss eval_loss
2025-09-21 11:21:20 3 26.5189
2025-09-21 11:21:26 6 25.7779
2nd phase with MTP
time step loss eval_loss
2025-09-25 22:34:46 140
2025-09-25 22:37:01 175 20.5087
2025-09-25 22:37:16 175 2.7290408611297607
2025-09-25 22:48:17 350 10.3879
2025-09-25 22:48:31 350 1.9787099361419678
2025-09-25 22:59:39 525 8.9108
2025-09-25 22:59:54 525 1.8240517377853394
2025-09-25 23:10:48 700 8.244
2025-09-25 23:11:03 700 1.7191474437713623
2025-09-25 23:11:04 700
Snippet of RL Training
Curriculum Update: max_new_tokens = 25
RL Training: 30%|███████████████████▊ | 300/1000 [04:30<12:59, 1.11s/it]
[RL Step 300] Loss=-0.0993 | Valid=1.000 | Lipinski=1.000 | Reward=0.469 | Entropy=1.423 | EntropyW=0.0100
Sample SELFIES: [=C] [C] [=C] [Branch2] [Branch1] [#C] [/C] [=C] [/C] [=Branch2] [Ring1] [Branch1] [=C] [Branch1] [C
Sample SMILES: C1C=C/C=C/C1=C(O)C(C2=CC=NC=C2)=CC=CC=C
Sample SELFIES: [=C] [C] [=C] [Branch2] [Branch1] [#C] [/C] [=C] [/C] [=Branch2] [Ring1] [Branch1] [=C] [Branch1] [C
Sample SMILES: C1C=C/C=C/C1=C(O)C(C2=CC=NC=C2)=CC=CC=C
RL Training: 35%|███████████████████████ | 350/1000 [05:39<14:51, 1.37s/it]
[RL Step 350] Loss=-0.2551 | Valid=1.000 | Lipinski=0.875 | Reward=0.382 | Entropy=1.295 | EntropyW=0.0100
Sample SELFIES: [=N+1] [Branch1] [C] [O-1] [C] [=C] [C] [=C] [Branch2] [Branch1] [=N] [S] [=Branch1] [C] [=O] [=Bran
Sample SMILES: [N+1]([O-1])C=CC=CS(=O)(=O)NC=CC=COCC(=O)C=CC=C
Sample SELFIES: [=N+1] [Branch1] [C] [O-1] [C] [=C] [C] [=C] [Branch2] [Branch1] [=N] [S] [=Branch1] [C] [=O] [=Bran
Sample SMILES: [N+1]([O-1])C=CC=CS(=O)(=O)NC=CC=COCC(=O)C=CC=C
Hardware it was trained on: Laptop with NVIDIA GeForce 930M GPU (2GB VRAM), RAM 12 GB, 2 cores Intel i3, SSD
🚀 Quick Start
- Clone this repository
- Make sure you have the requierements installed
- Configurable via
config.json - Run
python train-withmtp.py - Demo for generation with rendered mol image included in
demo_test_mtpresult.ipynb- For demo please extract the
pretrained.7zarchive
- For demo please extract the
- For testing the prototype PPO-KL RL fine-tuning, try running
train_ppokl_selfies.pyon the pretrained model (please make sure the model location is correct)
Tip: feel free to play around with the ChemQ3Model and its training loop/configs! The sample dataset is included so you can experiment with it~ especially if you have better compute than mine, feel free to share your results in discussion
To-Do
- Adjust FastChemTokenizer tokenizer on new data
- Experimenting with early architecture
- Write initial readme
- Upload backbone and MTP train code
- Demo training on 14K data (only 1 epoch, adding another on this data led to slight overfitting)
- Upload the warmup model
- Tidy up and upload JupyterNotebook(s) train/demo along with sample data
- [ongoing] Review, clean, and test codes
- Pretraining again after auditing/reviewing the base code
- Test RL code
- Train for 1000 steps for max token length = 64
- Upload RL-trained demo model
- Retrain MTP with fixed SELFIES tokenizer
- Retrain RL with fixed SELFIES tokenizer
- Update model on HF
- Update model and codes on GitHub
- Ablation studies
- Implement HF Automodel compatible modules if performance benefit(s) confirmed
- Complete pretraining on all ~3M dataset (when possible)
- Chunk I
- Chunk II
- Chunk III
- Chunk IV
- Publish complete pretraining on GitHub and HF (if compatible)
- Complete RL fine-tuning on verified rewards system.
📁 Project Structure
ChemMiniQ3-HoriFIE/
├── ChemQ3MTP.py # Custom model definition
|── train-withmtp.py # Main trainer for MTP with curriculum training combining NTP with MTP
|── config.json # Configuration for model definition and training
|── FastChemTokenizer.py # FastChemTokenizer module
|── train_ppokl_selfies.py # Prototype PPO-KL RL training script
├── README.md
├── requirements.txt # I'd recommend making a conda env for this or you could try using different
versions and please note if you encounter a bug
└── selftok_core # FastChemTokenizer: SELFIES core used for this model, you can try _wtails
if you want to experiment
└── pretrained/
└── sample-e1/ # Pre-trained weights on sample 14k dataset, 1st epoch
└── sample-RL/
└── demo_test_mtpresult.ipynb # Demo script for generating SELFIES using pretrained model
└── log_train.txt # Pre-training console outputs on MTP train
└── data/ # 14k samples from combined dataset
🔧 Contributing
This project is a learning experiment — all contributions are welcome!
- 🧠 Have a better way to implement the methods?
- 📊 Want to add evaluation metrics?
- ✨ Found a bug? Please open an issue!
👉 Please:
- Keep changes minimal and focused.
- Add comments if you change core logic.
⚠️ Disclaimer
This is NOT a production model.
- Built during late-night prototyping sessions 🌙
- Not thoroughly validated or benchmarked due to compute constraint
- Some components are heuristic and unproven
- May crash, overfit, or generate nonsense (especially outside molecular data)
- I’m still learning PyTorch, attention mechanisms, and transformer internals
Use this code to learn and experiment — not to deploy.
📜 License
MIT
❤️ Acknowledgments
Based and Inspired by:
- https://github.com/KellerJordan/modded-nanogpt
- https://huggingface.co/docs/transformers/en/model_doc/t5gemma
- https://github.com/aspuru-guzik-group/selfies/
- https://github.com/lessw2020/Ranger21
- https://huggingface.co/gbyuvd/chemfie-gpt-experiment-1
- https://huggingface.co/gbyuvd/bionat-selfies-gen-tokenizer-wordlevel
- Old ChemZiRo-GPT experiment with adding RoPE, GQA, MTP, RMSProp to backbone GPT2 architecture
References
BibTeX
Qwen2
@misc{yang2024qwen2technicalreport,
title={Qwen2 Technical Report},
author={An Yang and Baosong Yang and Binyuan Hui and Bo Zheng and Bowen Yu and Chang Zhou and Chengpeng Li and Chengyuan Li and Dayiheng Liu and Fei Huang and Guanting Dong and Haoran Wei and Huan Lin and Jialong Tang and Jialin Wang and Jian Yang and Jianhong Tu and Jianwei Zhang and Jianxin Ma and Jianxin Yang and Jin Xu and Jingren Zhou and Jinze Bai and Jinzheng He and Junyang Lin and Kai Dang and Keming Lu and Keqin Chen and Kexin Yang and Mei Li and Mingfeng Xue and Na Ni and Pei Zhang and Peng Wang and Ru Peng and Rui Men and Ruize Gao and Runji Lin and Shijie Wang and Shuai Bai and Sinan Tan and Tianhang Zhu and Tianhao Li and Tianyu Liu and Wenbin Ge and Xiaodong Deng and Xiaohuan Zhou and Xingzhang Ren and Xinyu Zhang and Xipin Wei and Xuancheng Ren and Xuejing Liu and Yang Fan and Yang Yao and Yichang Zhang and Yu Wan and Yunfei Chu and Yuqiong Liu and Zeyu Cui and Zhenru Zhang and Zhifang Guo and Zhihao Fan},
year={2024},
eprint={2407.10671},
archivePrefix={arXiv},
primaryClass={cs.CL},
url={https://arxiv.org/abs/2407.10671},
}
COCONUTDB
@article{sorokina2021coconut,
title={COCONUT online: Collection of Open Natural Products database},
author={Sorokina, Maria and Merseburger, Peter and Rajan, Kohulan and Yirik, Mehmet Aziz and Steinbeck, Christoph},
journal={Journal of Cheminformatics},
volume={13},
number={1},
pages={2},
year={2021},
doi={10.1186/s13321-020-00478-9}
}
ChemBL34
@article{zdrazil2023chembl,
title={The ChEMBL Database in 2023: a drug discovery platform spanning multiple bioactivity data types and time periods},
author={Zdrazil, Barbara and Felix, Eloy and Hunter, Fiona and Manners, Emma J and Blackshaw, James and Corbett, Sybilla and de Veij, Marleen and Ioannidis, Harris and Lopez, David Mendez and Mosquera, Juan F and Magarinos, Maria Paula and Bosc, Nicolas and Arcila, Ricardo and Kizil{\"o}ren, Tevfik and Gaulton, Anna and Bento, A Patr{\'i}cia and Adasme, Melissa F and Monecke, Peter and Landrum, Gregory A and Leach, Andrew R},
journal={Nucleic Acids Research},
year={2023},
volume={gkad1004},
doi={10.1093/nar/gkad1004}
}
@misc{chembl34,
title={ChemBL34},
year={2023},
doi={10.6019/CHEMBL.database.34}
}
SuperNatural3
@article{Gallo2023,
author = {Gallo, K and Kemmler, E and Goede, A and Becker, F and Dunkel, M and Preissner, R and Banerjee, P},
title = {{SuperNatural 3.0-a database of natural products and natural product-based derivatives}},
journal = {Nucleic Acids Research},
year = {2023},
month = jan,
day = {6},
volume = {51},
number = {D1},
pages = {D654-D659},
doi = {10.1093/nar/gkac1008}
}
Ranger21 Optimizer
@article{wright2021ranger21,
title={Ranger21: a synergistic deep learning optimizer},
author={Wright, Less and Demeure, Nestor},
year={2021},
journal={arXiv preprint arXiv:2106.13731},
}
- Downloads last month
- 22
