Upload 40 files
Browse files- datasets/adult.arff +0 -0
- datasets/bank-full.arff +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.001.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.002.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.003.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.004.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.005.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.006.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.007.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.008.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.009.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.010.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.011.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.012.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.013.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.014.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.015.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.016.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.017.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.018.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.019.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.020.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.021.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.022.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.023.png +0 -0
- figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.024.png +0 -0
- figures/adults_fooled.png +0 -0
- figures/adults_not_fooled.png +0 -0
- figures/bank_full_fooled.png +0 -0
- figures/bank_full_not_fooled.png +0 -0
- figures/data_adult_ep_10_bs_128_lr_0.001_al_0.5_dr_0.5_losses.png +0 -0
- figures/data_adult_ep_10_bs_128_lr_0.001_al_0.5_dr_0.5_pca.png +0 -0
- figures/data_bank-full_ep_10_bs_128_lr_0.001_al_0.2_dr_0.3_losses.png +0 -0
- figures/data_bank-full_ep_10_bs_128_lr_0.001_al_0.2_dr_0.3_pca.png +0 -0
- input_data/adult.arff +0 -0
- input_data/bank-full.arff +0 -0
- nt_exp.py +203 -0
- nt_gan.py +333 -0
- nt_gg.py +282 -0
- outputs/empty +1 -0
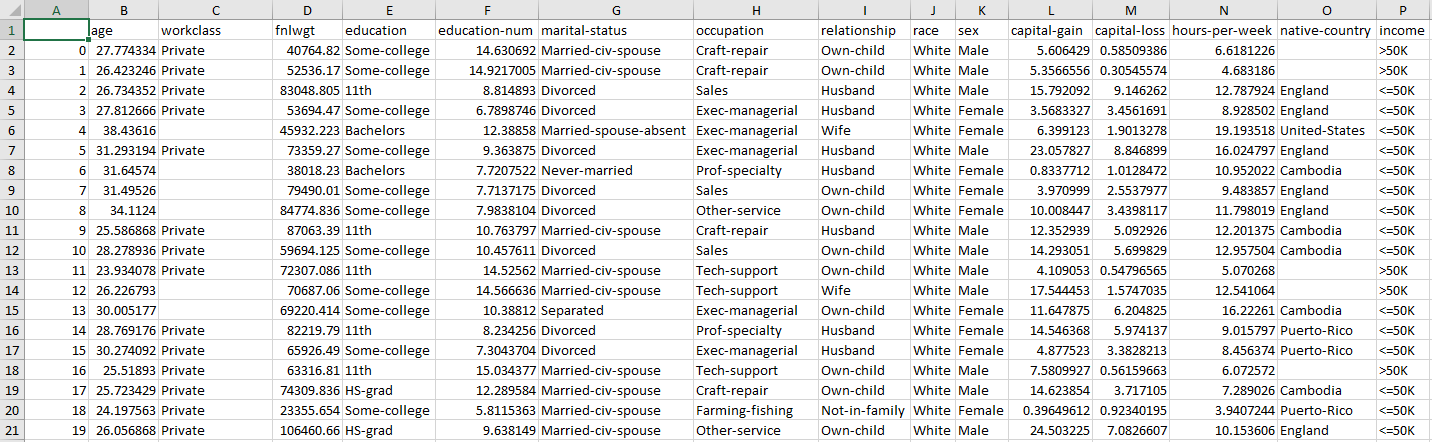
datasets/adult.arff
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
datasets/bank-full.arff
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.001.png
ADDED
|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.002.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.003.png
ADDED
|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.004.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.005.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.006.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.007.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.008.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.009.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.010.png
ADDED
|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.011.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.012.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.013.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.014.png
ADDED
|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.015.png
ADDED
|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.016.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.017.png
ADDED
|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.018.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.019.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.020.png
ADDED
|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.021.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.022.png
ADDED
|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.023.png
ADDED

|
figures/Aspose.Words.36be2542-1776-4b1c-8010-360ae82480ae.024.png
ADDED

|
figures/adults_fooled.png
ADDED
|
figures/adults_not_fooled.png
ADDED

|
figures/bank_full_fooled.png
ADDED

|
figures/bank_full_not_fooled.png
ADDED

|
figures/data_adult_ep_10_bs_128_lr_0.001_al_0.5_dr_0.5_losses.png
ADDED

|
figures/data_adult_ep_10_bs_128_lr_0.001_al_0.5_dr_0.5_pca.png
ADDED

|
figures/data_bank-full_ep_10_bs_128_lr_0.001_al_0.2_dr_0.3_losses.png
ADDED

|
figures/data_bank-full_ep_10_bs_128_lr_0.001_al_0.2_dr_0.3_pca.png
ADDED

|
input_data/adult.arff
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
input_data/bank-full.arff
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
nt_exp.py
ADDED
|
@@ -0,0 +1,203 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
|
| 3 |
+
import numpy as np
|
| 4 |
+
import pandas as pd
|
| 5 |
+
from scipy.io import arff
|
| 6 |
+
from sklearn import preprocessing
|
| 7 |
+
from sklearn.model_selection import train_test_split
|
| 8 |
+
|
| 9 |
+
from nt_gan import GAN
|
| 10 |
+
from nt_gg import GG
|
| 11 |
+
|
| 12 |
+
dataset_directory = 'datasets'
|
| 13 |
+
saved_models_path = 'outputs'
|
| 14 |
+
|
| 15 |
+
|
| 16 |
+
def prepare_architecture(arff_data_path):
|
| 17 |
+
"""
|
| 18 |
+
This function create the architecture of the GAN network.
|
| 19 |
+
The generator and the discriminator are created and then combined into the GAN model
|
| 20 |
+
:param arff_data_path: data path for the arff file
|
| 21 |
+
:return: a dictionary with all the relevant variables for the next stages
|
| 22 |
+
"""
|
| 23 |
+
data, meta_data = arff.loadarff(arff_data_path) # This function reads arff file into tuple of data and its meta.
|
| 24 |
+
df = pd.DataFrame(data)
|
| 25 |
+
columns = df.columns
|
| 26 |
+
transformed_data, x, x_scaled, meta_data_rev, min_max_scaler = create_scaled_data(df, meta_data)
|
| 27 |
+
|
| 28 |
+
number_of_features = len(transformed_data.columns) # Define the GAN and training parameters
|
| 29 |
+
|
| 30 |
+
return x_scaled, meta_data_rev, columns, min_max_scaler, number_of_features
|
| 31 |
+
|
| 32 |
+
|
| 33 |
+
def create_scaled_data(df, meta_data):
|
| 34 |
+
"""
|
| 35 |
+
|
| 36 |
+
:param df:
|
| 37 |
+
:param meta_data:
|
| 38 |
+
:return:
|
| 39 |
+
"""
|
| 40 |
+
meta_data_dict = {k: {a.replace(' ', ''): b + 1 for b, a in enumerate(v.values)} for k, v in
|
| 41 |
+
meta_data._attributes.items() if
|
| 42 |
+
v.type_name != 'numeric'} # Starts from one and not zero because one is for Nan values
|
| 43 |
+
meta_data_rev = {k: {b + 1: a.replace(' ', '') for b, a in enumerate(v.values)} for k, v in
|
| 44 |
+
meta_data._attributes.items() if
|
| 45 |
+
v.type_name != 'numeric'} # Starts from one and not zero because one is for Nan values
|
| 46 |
+
transformed_data = df.copy()
|
| 47 |
+
for col in df.columns:
|
| 48 |
+
if col in meta_data_dict:
|
| 49 |
+
# Sometimes the values can not be found in the meta data, so we treat these values as Nan
|
| 50 |
+
transformed_data[col] = transformed_data[col].apply(
|
| 51 |
+
lambda x: meta_data_dict[col][str(x).split('\'')[1]] if str(x).split('\'')[1] in meta_data_dict[
|
| 52 |
+
col] else 0)
|
| 53 |
+
x = transformed_data.values # returns a numpy array
|
| 54 |
+
min_max_scaler = preprocessing.MinMaxScaler()
|
| 55 |
+
x_scaled = min_max_scaler.fit_transform(x)
|
| 56 |
+
return transformed_data, x, x_scaled, meta_data_rev, min_max_scaler
|
| 57 |
+
|
| 58 |
+
|
| 59 |
+
def re_scaled_data(data, columns, meta_data_rev, min_max_scaler):
|
| 60 |
+
"""
|
| 61 |
+
This function re-scaled the fake data to the original format.
|
| 62 |
+
:param data: the data we want to re scaled
|
| 63 |
+
:param columns:
|
| 64 |
+
:param meta_data_rev:
|
| 65 |
+
:return:
|
| 66 |
+
"""
|
| 67 |
+
data_inv = min_max_scaler.inverse_transform(data)
|
| 68 |
+
df = pd.DataFrame(data_inv, columns=columns)
|
| 69 |
+
transformed_data = df.copy()
|
| 70 |
+
for col in transformed_data.columns:
|
| 71 |
+
if col in meta_data_rev:
|
| 72 |
+
# Sometimes the values can not be found in the meta data, so we treat these values as Nan
|
| 73 |
+
transformed_data[col] = transformed_data[col].apply(
|
| 74 |
+
lambda x: meta_data_rev[col][int(round(x))] if int(round(x)) in meta_data_rev[
|
| 75 |
+
col] else np.nan)
|
| 76 |
+
return transformed_data
|
| 77 |
+
|
| 78 |
+
|
| 79 |
+
def first_question():
|
| 80 |
+
"""
|
| 81 |
+
This function answers the first question
|
| 82 |
+
:return:
|
| 83 |
+
"""
|
| 84 |
+
to_plot_losses = True
|
| 85 |
+
results_output = os.path.join(saved_models_path, f'question_one_results.csv')
|
| 86 |
+
results = {'dataset': [], 'lr': [], 'ep': [], 'bs': [], 'alpha': [], 'dropout': [], 'gen_loss': [], 'dis_loss': [],
|
| 87 |
+
'activation': [], 'fooled_len': [], 'not_fooled_len': [], 'mean_min_distance_fooled': [],
|
| 88 |
+
'mean_min_distance_not_fooled': [], 'mean_min_distance_gap': []}
|
| 89 |
+
# w1 * (MMDF + MMDNF) - w3 * (MMDG) + w2 * (NFL/ 100)
|
| 90 |
+
# MMDG = MMDNF - MMDF
|
| 91 |
+
# data_name = ["adult", "bank-full"]
|
| 92 |
+
# learning_rate = [0.01, 0.001, 0.0001]
|
| 93 |
+
# epochs = [5, 10, 15]
|
| 94 |
+
# batch_size = [64, 128, 1024]
|
| 95 |
+
# alpha_relu = [0.2, 0.5]
|
| 96 |
+
# dropout = [0.3, 0.5]
|
| 97 |
+
data_name = ["adult"]
|
| 98 |
+
learning_rate = [0.001]
|
| 99 |
+
epochs = [10]
|
| 100 |
+
batch_size = [128]
|
| 101 |
+
alpha_relu = [0.5]
|
| 102 |
+
dropout = [0.5]
|
| 103 |
+
loss = 'binary_crossentropy'
|
| 104 |
+
activation = 'sigmoid'
|
| 105 |
+
|
| 106 |
+
for data in data_name:
|
| 107 |
+
for lr in learning_rate:
|
| 108 |
+
for ep in epochs:
|
| 109 |
+
for bs in batch_size:
|
| 110 |
+
for al in alpha_relu:
|
| 111 |
+
for dr in dropout:
|
| 112 |
+
arff_data_path = f'./datasets/{data}.arff'
|
| 113 |
+
model_name = f'data_{data}_ep_{ep}_bs_{bs}_lr_{lr}_al_{al}_dr_{dr}'
|
| 114 |
+
pca_output = os.path.join(saved_models_path, f'{model_name}_pca.png')
|
| 115 |
+
fooled_output = os.path.join(saved_models_path, f'{model_name}_fooled.csv')
|
| 116 |
+
not_fooled_output = os.path.join(saved_models_path, f'{model_name}_not_fooled.csv')
|
| 117 |
+
|
| 118 |
+
x_scaled, meta_data_rev, columns, min_max_scaler, number_of_features = prepare_architecture(
|
| 119 |
+
arff_data_path)
|
| 120 |
+
gan_obj = GAN(number_of_features=number_of_features, saved_models_path=saved_models_path,
|
| 121 |
+
learning_rate=lr, alpha_relu=al, dropout=dr,
|
| 122 |
+
loss=loss, activation=activation)
|
| 123 |
+
gen_loss, dis_loss = gan_obj.train(scaled_data=x_scaled, epochs=ep, batch_size=bs,
|
| 124 |
+
to_plot_losses=to_plot_losses, model_name=model_name)
|
| 125 |
+
dis_fooled_scaled, dis_not_fooled_scaled, mean_min_distance_fooled, mean_min_distance_not_fooled = gan_obj.test(
|
| 126 |
+
scaled_data=x_scaled, sample_num=100, pca_output=pca_output)
|
| 127 |
+
dis_fooled = re_scaled_data(data=dis_fooled_scaled, columns=columns,
|
| 128 |
+
meta_data_rev=meta_data_rev,
|
| 129 |
+
min_max_scaler=min_max_scaler)
|
| 130 |
+
dis_fooled.to_csv(fooled_output)
|
| 131 |
+
dis_not_fooled = re_scaled_data(data=dis_not_fooled_scaled, columns=columns,
|
| 132 |
+
meta_data_rev=meta_data_rev,
|
| 133 |
+
min_max_scaler=min_max_scaler)
|
| 134 |
+
dis_not_fooled.to_csv(not_fooled_output)
|
| 135 |
+
results['dataset'].append(data)
|
| 136 |
+
results['lr'].append(lr)
|
| 137 |
+
results['ep'].append(ep)
|
| 138 |
+
results['bs'].append(bs)
|
| 139 |
+
results['alpha'].append(al)
|
| 140 |
+
results['dropout'].append(dr)
|
| 141 |
+
results['gen_loss'].append(gen_loss)
|
| 142 |
+
results['dis_loss'].append(dis_loss)
|
| 143 |
+
results['activation'].append(activation)
|
| 144 |
+
results['fooled_len'].append(len(dis_fooled_scaled))
|
| 145 |
+
results['not_fooled_len'].append(len(dis_not_fooled_scaled))
|
| 146 |
+
results['mean_min_distance_fooled'].append(mean_min_distance_fooled)
|
| 147 |
+
results['mean_min_distance_not_fooled'].append(mean_min_distance_not_fooled)
|
| 148 |
+
results['mean_min_distance_gap'].append(mean_min_distance_not_fooled-mean_min_distance_fooled)
|
| 149 |
+
results_df = pd.DataFrame.from_dict(results)
|
| 150 |
+
results_df.to_csv(results_output, index=False)
|
| 151 |
+
|
| 152 |
+
|
| 153 |
+
def second_question():
|
| 154 |
+
|
| 155 |
+
data_name = ["adult", "bank-full"]
|
| 156 |
+
learning_rate = [0.001]
|
| 157 |
+
epochs = [10]
|
| 158 |
+
batch_size = [128]
|
| 159 |
+
alpha_relu = [0.2]
|
| 160 |
+
dropout = [0.3]
|
| 161 |
+
results = {'dataset': [], 'lr': [], 'ep': [], 'bs': [], 'alpha': [], 'dropout': [], 'gen_loss': [], 'proba_error': []}
|
| 162 |
+
combs = len(data_name) * len(learning_rate) * len(epochs) * len(batch_size) * len(alpha_relu) * len(dropout)
|
| 163 |
+
i = 1
|
| 164 |
+
for data in data_name:
|
| 165 |
+
for lr in learning_rate:
|
| 166 |
+
for ep in epochs:
|
| 167 |
+
for bs in batch_size:
|
| 168 |
+
for al in alpha_relu:
|
| 169 |
+
for dr in dropout:
|
| 170 |
+
print(f'Running combination {i}/{combs}')
|
| 171 |
+
data_path = f'./datasets/{data}.arff'
|
| 172 |
+
model_name = f'data_{data}_ep_{ep}_bs_{bs}_lr_{lr}_part2'
|
| 173 |
+
x_scaled, meta_data_rev, cols, min_max_scaler, feature_num = prepare_architecture(data_path)
|
| 174 |
+
general_generator = GG(feature_num, saved_models_path, lr, dr, al)
|
| 175 |
+
x_train, x_test, y_train, y_test = train_test_split(x_scaled[:, :-1], x_scaled[:, -1], test_size=0.1)
|
| 176 |
+
general_generator.train_gg(x_train, y_train, ep, bs, model_name, data, saved_models_path, True)
|
| 177 |
+
error = general_generator.get_error()
|
| 178 |
+
results['dataset'].append(data)
|
| 179 |
+
results['lr'].append(lr)
|
| 180 |
+
results['ep'].append(ep)
|
| 181 |
+
results['bs'].append(bs)
|
| 182 |
+
results['alpha'].append(al)
|
| 183 |
+
results['dropout'].append(dr)
|
| 184 |
+
results['gen_loss'].append(general_generator.losses['gen_loss'][-1])
|
| 185 |
+
results['proba_error'].append(error.mean())
|
| 186 |
+
i += 1
|
| 187 |
+
# Test set performance
|
| 188 |
+
general_generator.plot_discriminator_results(x_test, y_test, data, saved_models_path)
|
| 189 |
+
general_generator.plot_generator_results(data, saved_models_path)
|
| 190 |
+
|
| 191 |
+
results_output = os.path.join(saved_models_path, f'question_two_results.csv')
|
| 192 |
+
results_df = pd.DataFrame.from_dict(results)
|
| 193 |
+
# results_df.to_csv(results_output, index=False)
|
| 194 |
+
|
| 195 |
+
|
| 196 |
+
|
| 197 |
+
def main():
|
| 198 |
+
# first_question()
|
| 199 |
+
second_question()
|
| 200 |
+
|
| 201 |
+
|
| 202 |
+
if __name__ == '__main__':
|
| 203 |
+
main()
|
nt_gan.py
ADDED
|
@@ -0,0 +1,333 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
from itertools import compress
|
| 3 |
+
|
| 4 |
+
import matplotlib.pyplot as plt
|
| 5 |
+
import numpy as np
|
| 6 |
+
import pandas as pd
|
| 7 |
+
from keras.layers import Dense, Dropout, LeakyReLU
|
| 8 |
+
from keras.models import Sequential
|
| 9 |
+
from keras.optimizers import Adam
|
| 10 |
+
from numpy.random import randn
|
| 11 |
+
from sklearn.decomposition import PCA
|
| 12 |
+
from tqdm import tqdm
|
| 13 |
+
|
| 14 |
+
|
| 15 |
+
class GAN(object):
|
| 16 |
+
def __init__(self, number_of_features, saved_models_path, learning_rate, alpha_relu, dropout, loss, activation):
|
| 17 |
+
"""
|
| 18 |
+
A constructor for the GAN class
|
| 19 |
+
:param number_of_features: number of features
|
| 20 |
+
:param saved_models_path: the output folder path
|
| 21 |
+
"""
|
| 22 |
+
self.saved_models_path = saved_models_path
|
| 23 |
+
self.number_of_features = number_of_features
|
| 24 |
+
|
| 25 |
+
self.generator_model = None
|
| 26 |
+
self.noise_dim = None
|
| 27 |
+
self.discriminator_model = None
|
| 28 |
+
self.learning_rate = learning_rate
|
| 29 |
+
self.gan_model = None
|
| 30 |
+
self.activation = activation
|
| 31 |
+
self.alpha_relu = alpha_relu
|
| 32 |
+
self.loss = loss
|
| 33 |
+
self.dropout = dropout
|
| 34 |
+
self.number_of_features = number_of_features
|
| 35 |
+
|
| 36 |
+
self.build_generator() # build the generator
|
| 37 |
+
self.build_discriminator() # build the discriminator
|
| 38 |
+
self.build_gan() # build the GAN
|
| 39 |
+
|
| 40 |
+
def build_generator(self):
|
| 41 |
+
"""
|
| 42 |
+
This function creates the generator model
|
| 43 |
+
:return:
|
| 44 |
+
"""
|
| 45 |
+
noise_size = int(self.number_of_features / 2)
|
| 46 |
+
self.noise_dim = (noise_size,) # size of the noise space
|
| 47 |
+
|
| 48 |
+
self.generator_model = Sequential()
|
| 49 |
+
self.generator_model.add(Dense(int(self.number_of_features * 2), input_shape=self.noise_dim))
|
| 50 |
+
self.generator_model.add(LeakyReLU(alpha=self.alpha_relu))
|
| 51 |
+
|
| 52 |
+
self.generator_model.add(Dense(int(self.number_of_features * 4)))
|
| 53 |
+
self.generator_model.add(LeakyReLU(alpha=self.alpha_relu))
|
| 54 |
+
self.generator_model.add(Dropout(self.dropout))
|
| 55 |
+
|
| 56 |
+
self.generator_model.add(Dense(int(self.number_of_features * 2)))
|
| 57 |
+
self.generator_model.add(LeakyReLU(alpha=self.alpha_relu))
|
| 58 |
+
self.generator_model.add(Dropout(self.dropout))
|
| 59 |
+
|
| 60 |
+
# Compile it
|
| 61 |
+
self.generator_model.add(Dense(self.number_of_features, activation=self.activation))
|
| 62 |
+
self.generator_model.summary()
|
| 63 |
+
|
| 64 |
+
def build_discriminator(self):
|
| 65 |
+
"""
|
| 66 |
+
Create discriminator model
|
| 67 |
+
:return:
|
| 68 |
+
"""
|
| 69 |
+
self.discriminator_model = Sequential()
|
| 70 |
+
|
| 71 |
+
self.discriminator_model.add(Dense(self.number_of_features * 2, input_shape=(self.number_of_features,)))
|
| 72 |
+
self.discriminator_model.add(LeakyReLU(alpha=self.alpha_relu))
|
| 73 |
+
|
| 74 |
+
self.discriminator_model.add(Dense(self.number_of_features * 4))
|
| 75 |
+
self.discriminator_model.add(LeakyReLU(alpha=self.alpha_relu))
|
| 76 |
+
self.discriminator_model.add(Dropout(self.dropout))
|
| 77 |
+
|
| 78 |
+
self.discriminator_model.add(Dense(self.number_of_features * 2))
|
| 79 |
+
self.discriminator_model.add(LeakyReLU(alpha=self.alpha_relu))
|
| 80 |
+
self.discriminator_model.add(Dropout(self.dropout))
|
| 81 |
+
|
| 82 |
+
# Compile it
|
| 83 |
+
self.discriminator_model.add(Dense(1, activation=self.activation))
|
| 84 |
+
optimizer = Adam(lr=self.learning_rate)
|
| 85 |
+
self.discriminator_model.compile(loss=self.loss, optimizer=optimizer)
|
| 86 |
+
self.discriminator_model.summary()
|
| 87 |
+
|
| 88 |
+
def build_gan(self):
|
| 89 |
+
"""
|
| 90 |
+
Create the GAN network
|
| 91 |
+
:return: the GAN model object
|
| 92 |
+
"""
|
| 93 |
+
self.gan_model = Sequential()
|
| 94 |
+
self.discriminator_model.trainable = False
|
| 95 |
+
|
| 96 |
+
# The following lines connect the generator and discriminator models to the GAN.
|
| 97 |
+
self.gan_model.add(self.generator_model)
|
| 98 |
+
self.gan_model.add(self.discriminator_model)
|
| 99 |
+
|
| 100 |
+
# Compile it
|
| 101 |
+
optimizer = Adam(lr=self.learning_rate)
|
| 102 |
+
self.gan_model.compile(loss=self.loss, optimizer=optimizer)
|
| 103 |
+
|
| 104 |
+
return self.gan_model
|
| 105 |
+
|
| 106 |
+
def train(self, scaled_data, epochs, batch_size, to_plot_losses, model_name):
|
| 107 |
+
"""
|
| 108 |
+
This function trains the generator and discriminator outputs
|
| 109 |
+
:param model_name:
|
| 110 |
+
:param to_plot_losses: whether or not to plot history
|
| 111 |
+
:param scaled_data: the data after min max scaling
|
| 112 |
+
:param epochs: number of epochs
|
| 113 |
+
:param batch_size: the batch size
|
| 114 |
+
:return: losses_list: returns the losses dictionary the generator or discriminator outputs
|
| 115 |
+
"""
|
| 116 |
+
dis_output, gen_output, prev_output = self.check_for_existed_output(model_name)
|
| 117 |
+
if prev_output:
|
| 118 |
+
return -1, -1
|
| 119 |
+
|
| 120 |
+
losses_output = os.path.join(self.saved_models_path, f'{model_name}_losses.png')
|
| 121 |
+
discriminator_loss = []
|
| 122 |
+
generator_loss = []
|
| 123 |
+
|
| 124 |
+
# We need to use half of the batch size for the fake data and half for the real one
|
| 125 |
+
half_batch_size = int(batch_size / 2)
|
| 126 |
+
iterations = int(len(scaled_data) / half_batch_size)
|
| 127 |
+
iterations = iterations + 1 if len(scaled_data) % batch_size != 0 else iterations
|
| 128 |
+
|
| 129 |
+
for epoch in range(1, epochs + 1): # iterates over the epochs
|
| 130 |
+
np.random.shuffle(scaled_data)
|
| 131 |
+
p_bar = tqdm(range(iterations), ascii=True)
|
| 132 |
+
for iteration in p_bar:
|
| 133 |
+
dis_loss, gen_loss = self.train_models(batch_size=batch_size, half_batch_size=half_batch_size,
|
| 134 |
+
index=iteration, scaled_data=scaled_data)
|
| 135 |
+
discriminator_loss.append(dis_loss)
|
| 136 |
+
generator_loss.append(gen_loss)
|
| 137 |
+
p_bar.set_description(
|
| 138 |
+
f"Epoch ({epoch}/{epochs}) | DISCRIMINATOR LOSS: {dis_loss:.2f} | GENERATOR LOSS: {gen_loss:.2f} |")
|
| 139 |
+
|
| 140 |
+
# Save weights for future use
|
| 141 |
+
self.discriminator_model.save_weights(dis_output)
|
| 142 |
+
self.generator_model.save_weights(gen_output)
|
| 143 |
+
|
| 144 |
+
# Plot losses
|
| 145 |
+
if to_plot_losses:
|
| 146 |
+
self.plot_losses(discriminator_loss=discriminator_loss, generator_loss=generator_loss,
|
| 147 |
+
losses_output=losses_output)
|
| 148 |
+
|
| 149 |
+
return generator_loss[-1], discriminator_loss[-1]
|
| 150 |
+
|
| 151 |
+
def check_for_existed_output(self, model_name) -> (str, str, bool):
|
| 152 |
+
"""
|
| 153 |
+
This function checks for existed output
|
| 154 |
+
:param model_name: model's name
|
| 155 |
+
:return:
|
| 156 |
+
"""
|
| 157 |
+
prev_output = False
|
| 158 |
+
dis_output = os.path.join(self.saved_models_path, f'{model_name}_dis_weights.h5')
|
| 159 |
+
gen_output = os.path.join(self.saved_models_path, f'{model_name}_gen_weights.h5')
|
| 160 |
+
if os.path.exists(dis_output) and os.path.exists(gen_output):
|
| 161 |
+
print("The model was trained in the past")
|
| 162 |
+
self.discriminator_model.load_weights(dis_output)
|
| 163 |
+
self.generator_model.load_weights(gen_output)
|
| 164 |
+
prev_output = True
|
| 165 |
+
return dis_output, gen_output, prev_output
|
| 166 |
+
|
| 167 |
+
def train_models(self, batch_size, half_batch_size, index, scaled_data):
|
| 168 |
+
"""
|
| 169 |
+
This function trains the discriminator and the generator
|
| 170 |
+
:param batch_size: batch size
|
| 171 |
+
:param half_batch_size: half of the batch size
|
| 172 |
+
:param index:
|
| 173 |
+
:param scaled_data:
|
| 174 |
+
:return:
|
| 175 |
+
"""
|
| 176 |
+
self.discriminator_model.trainable = True
|
| 177 |
+
|
| 178 |
+
# Create a batch of real data and train the model
|
| 179 |
+
x_real, y_real = self.get_real_samples(data=scaled_data, batch_size=half_batch_size, index=index)
|
| 180 |
+
d_real_loss = self.discriminator_model.train_on_batch(x_real, y_real)
|
| 181 |
+
|
| 182 |
+
# Create a batch of fake data and train the model
|
| 183 |
+
x_fake, y_fake = self.create_fake_samples(batch_size=half_batch_size)
|
| 184 |
+
d_fake_loss = self.discriminator_model.train_on_batch(x_fake, y_fake)
|
| 185 |
+
|
| 186 |
+
avg_dis_loss = 0.5 * (d_real_loss + d_fake_loss)
|
| 187 |
+
|
| 188 |
+
# Create noise for the generator model
|
| 189 |
+
noise = randn(self.noise_dim[0] * batch_size).reshape((batch_size, self.noise_dim[0]))
|
| 190 |
+
|
| 191 |
+
self.discriminator_model.trainable = False
|
| 192 |
+
gen_loss = self.gan_model.train_on_batch(noise, np.ones((batch_size, 1)))
|
| 193 |
+
|
| 194 |
+
return avg_dis_loss, gen_loss
|
| 195 |
+
|
| 196 |
+
@staticmethod
|
| 197 |
+
def get_real_samples(data, batch_size, index):
|
| 198 |
+
"""
|
| 199 |
+
Generate batch_size of real samples with class labels
|
| 200 |
+
:param data: the original data
|
| 201 |
+
:param batch_size: batch size
|
| 202 |
+
:param index: the index of the batch
|
| 203 |
+
:return: x: real samples, y: labels
|
| 204 |
+
"""
|
| 205 |
+
start_index = batch_size * index
|
| 206 |
+
end_index = start_index + batch_size
|
| 207 |
+
x = data[start_index: end_index]
|
| 208 |
+
|
| 209 |
+
return x, np.ones((len(x), 1))
|
| 210 |
+
|
| 211 |
+
def create_fake_samples(self, batch_size):
|
| 212 |
+
"""
|
| 213 |
+
Use the generator to generate n fake examples, with class labels
|
| 214 |
+
:param batch_size: batch size
|
| 215 |
+
:return:
|
| 216 |
+
"""
|
| 217 |
+
noise = randn(self.noise_dim[0] * batch_size).reshape((batch_size, self.noise_dim[0]))
|
| 218 |
+
x = self.generator_model.predict(noise) # create fake samples using the generator
|
| 219 |
+
|
| 220 |
+
return x, np.zeros((len(x), 1))
|
| 221 |
+
|
| 222 |
+
@staticmethod
|
| 223 |
+
def plot_losses(discriminator_loss, generator_loss, losses_output):
|
| 224 |
+
"""
|
| 225 |
+
Plot training loss values
|
| 226 |
+
:param generator_loss:
|
| 227 |
+
:param discriminator_loss:
|
| 228 |
+
:param losses_output:
|
| 229 |
+
:return:
|
| 230 |
+
"""
|
| 231 |
+
plt.plot(discriminator_loss)
|
| 232 |
+
plt.plot(generator_loss)
|
| 233 |
+
plt.xlabel('Iteration')
|
| 234 |
+
plt.ylabel('Loss')
|
| 235 |
+
plt.title('Discriminator and Generator Losses')
|
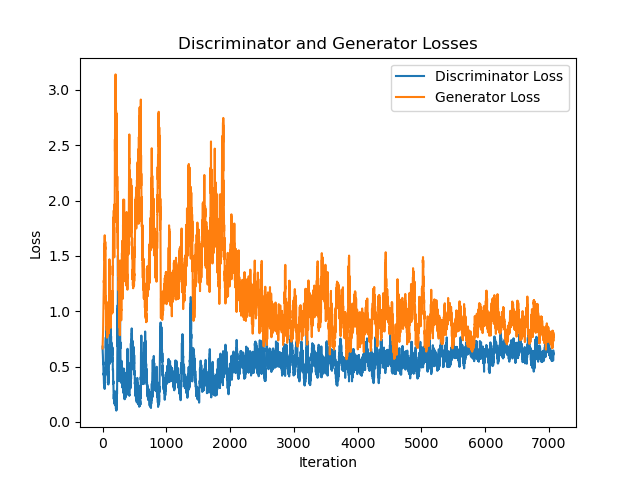
| 236 |
+
plt.legend(['Discriminator Loss', 'Generator Loss'])
|
| 237 |
+
plt.savefig(losses_output)
|
| 238 |
+
|
| 239 |
+
@staticmethod
|
| 240 |
+
def return_minimum_euclidean_distance(scaled_data, x):
|
| 241 |
+
"""
|
| 242 |
+
This function returns the
|
| 243 |
+
:param scaled_data: the original data
|
| 244 |
+
:param x: a record we want to compare with
|
| 245 |
+
:return: the minimum distance and the index of the minimum value
|
| 246 |
+
"""
|
| 247 |
+
s = np.power(np.power((scaled_data - np.array(x)), 2).sum(1), 0.5)
|
| 248 |
+
return pd.Series([s[s.argmin()], s.argmin()])
|
| 249 |
+
|
| 250 |
+
def test(self, scaled_data, sample_num, pca_output):
|
| 251 |
+
"""
|
| 252 |
+
This function tests the model
|
| 253 |
+
:param scaled_data: the original scaled data
|
| 254 |
+
:param sample_num: number of samples to generate
|
| 255 |
+
:param pca_output: the output of PCA
|
| 256 |
+
:return:
|
| 257 |
+
"""
|
| 258 |
+
x_fake, y_fake = self.create_fake_samples(batch_size=sample_num)
|
| 259 |
+
fake_pred = self.discriminator_model.predict(x_fake)
|
| 260 |
+
|
| 261 |
+
# Filter data to different matrices
|
| 262 |
+
dis_fooled_scaled = np.asarray(list(compress(x_fake, fake_pred > 0.5)))
|
| 263 |
+
dis_not_fooled_scaled = np.asarray(list(compress(x_fake, fake_pred <= 0.5)))
|
| 264 |
+
|
| 265 |
+
# ------------- Euclidean -------------
|
| 266 |
+
mean_min_distance_fooled, mean_min_distance_not_fooled = (-1, -1)
|
| 267 |
+
if len(dis_fooled_scaled) > 0 and len(dis_not_fooled_scaled) > 0:
|
| 268 |
+
mean_min_distance_fooled = self.get_mean_distance_score(scaled_data, dis_fooled_scaled)
|
| 269 |
+
print(f'The mean minimum distance for fooled samples is {mean_min_distance_fooled}')
|
| 270 |
+
mean_min_distance_not_fooled = self.get_mean_distance_score(scaled_data, dis_not_fooled_scaled)
|
| 271 |
+
print(f'The mean minimum distance for not fooled samples is {mean_min_distance_not_fooled}')
|
| 272 |
+
else:
|
| 273 |
+
print(f'The fooled xor the not Fooled data frames is empty')
|
| 274 |
+
|
| 275 |
+
# ------------- PCA --------------
|
| 276 |
+
data_pca_df = self.get_pca_df(scaled_data, 'original')
|
| 277 |
+
dis_fooled_pca_df = self.get_pca_df(dis_fooled_scaled, 'fooled')
|
| 278 |
+
dis_not_fooled_pca_df = self.get_pca_df(dis_not_fooled_scaled, 'not fooled')
|
| 279 |
+
pca_frames = [data_pca_df, dis_fooled_pca_df, dis_not_fooled_pca_df]
|
| 280 |
+
pca_result = pd.concat(pca_frames)
|
| 281 |
+
self.plot_pca(pca_result, pca_output)
|
| 282 |
+
|
| 283 |
+
return dis_fooled_scaled, dis_not_fooled_scaled, mean_min_distance_fooled, mean_min_distance_not_fooled
|
| 284 |
+
|
| 285 |
+
def get_mean_distance_score(self, scaled_data, dis_scaled):
|
| 286 |
+
"""
|
| 287 |
+
This function returns the mean distance score for the given dataframe
|
| 288 |
+
:param scaled_data: the original data
|
| 289 |
+
:param dis_scaled: a dataframe
|
| 290 |
+
:return:
|
| 291 |
+
"""
|
| 292 |
+
dis_fooled_scaled_ecu = pd.DataFrame(dis_scaled)
|
| 293 |
+
dis_fooled_scaled_ecu[['min_distance', 'similar_i']] = dis_fooled_scaled_ecu.apply(
|
| 294 |
+
lambda x: self.return_minimum_euclidean_distance(scaled_data, x), axis=1)
|
| 295 |
+
mean_min_distance_fooled = dis_fooled_scaled_ecu['min_distance'].mean()
|
| 296 |
+
return mean_min_distance_fooled
|
| 297 |
+
|
| 298 |
+
@staticmethod
|
| 299 |
+
def plot_pca(pca_result, pca_output):
|
| 300 |
+
"""
|
| 301 |
+
This function plots the PCA figure
|
| 302 |
+
:param pca_result: dataframe with all the results
|
| 303 |
+
:param pca_output: output path
|
| 304 |
+
:return:
|
| 305 |
+
"""
|
| 306 |
+
fig = plt.figure(figsize=(8, 8))
|
| 307 |
+
ax = fig.add_subplot(1, 1, 1)
|
| 308 |
+
ax.set_xlabel('Principal Component 1', fontsize=15)
|
| 309 |
+
ax.set_ylabel('Principal Component 2', fontsize=15)
|
| 310 |
+
ax.set_title('PCA With Two Components', fontsize=20)
|
| 311 |
+
targets = ['original', 'fooled', 'not fooled']
|
| 312 |
+
colors = ['r', 'g', 'b']
|
| 313 |
+
for target, color in zip(targets, colors):
|
| 314 |
+
indices_to_keep = pca_result['name'] == target
|
| 315 |
+
ax.scatter(pca_result.loc[indices_to_keep, 'comp1'], pca_result.loc[indices_to_keep, 'comp2'],
|
| 316 |
+
c=color, s=50)
|
| 317 |
+
ax.legend(targets)
|
| 318 |
+
ax.grid()
|
| 319 |
+
plt.savefig(pca_output)
|
| 320 |
+
|
| 321 |
+
@staticmethod
|
| 322 |
+
def get_pca_df(scaled_data, data_name):
|
| 323 |
+
"""
|
| 324 |
+
This function creates the PCA dataframe
|
| 325 |
+
:param scaled_data: the original data
|
| 326 |
+
:param data_name: the name of the column
|
| 327 |
+
:return:
|
| 328 |
+
"""
|
| 329 |
+
pca = PCA(n_components=2)
|
| 330 |
+
principal_components = pca.fit_transform(scaled_data)
|
| 331 |
+
principal_df = pd.DataFrame(data=principal_components, columns=['comp1', 'comp2'])
|
| 332 |
+
principal_df['name'] = data_name
|
| 333 |
+
return principal_df
|
nt_gg.py
ADDED
|
@@ -0,0 +1,282 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import pickle
|
| 3 |
+
import matplotlib.pyplot as plt
|
| 4 |
+
import numpy as np
|
| 5 |
+
from keras.layers import Dense, Dropout, LeakyReLU
|
| 6 |
+
from keras.models import Sequential
|
| 7 |
+
from keras.optimizers import Adam
|
| 8 |
+
from numpy.random import randn
|
| 9 |
+
from sklearn.ensemble import RandomForestClassifier
|
| 10 |
+
from sklearn import metrics
|
| 11 |
+
from tqdm import tqdm
|
| 12 |
+
|
| 13 |
+
|
| 14 |
+
class GG(object):
|
| 15 |
+
|
| 16 |
+
def __init__(self, number_of_features, saved_models_path, learning_rate, dropout, alpha):
|
| 17 |
+
"""
|
| 18 |
+
The constructor for the General Generator class.
|
| 19 |
+
:param number_of_features: Number of features in the data. Used to determine the noise dimensions
|
| 20 |
+
:param saved_models_path: The folder where we save the models.
|
| 21 |
+
"""
|
| 22 |
+
self.saved_models_path = saved_models_path
|
| 23 |
+
self.number_of_features = number_of_features
|
| 24 |
+
|
| 25 |
+
self.generator_model = None
|
| 26 |
+
self.discriminator_model = RandomForestClassifier()
|
| 27 |
+
self.dropout = dropout
|
| 28 |
+
self.alpha = alpha
|
| 29 |
+
self.noise_dim = int(number_of_features / 2)
|
| 30 |
+
self.learning_rate = learning_rate
|
| 31 |
+
self.number_of_features = number_of_features
|
| 32 |
+
self.build_generator() # build the generator.
|
| 33 |
+
self.losses = {'gen_loss': [], 'dis_loss_pred': [], 'dis_loss_proba': []}
|
| 34 |
+
# self.results = {}
|
| 35 |
+
|
| 36 |
+
def build_generator(self):
|
| 37 |
+
"""
|
| 38 |
+
This function creates the generator model for the GG.
|
| 39 |
+
We used a fairly simple MLP architecture.
|
| 40 |
+
:return:
|
| 41 |
+
"""
|
| 42 |
+
|
| 43 |
+
self.generator_model = Sequential()
|
| 44 |
+
self.generator_model.add(Dense(int(self.number_of_features * 2), input_shape=(self.noise_dim + 1, )))
|
| 45 |
+
self.generator_model.add(LeakyReLU(alpha=self.alpha))
|
| 46 |
+
|
| 47 |
+
self.generator_model.add(Dense(int(self.number_of_features * 4)))
|
| 48 |
+
self.generator_model.add(LeakyReLU(alpha=self.alpha))
|
| 49 |
+
self.generator_model.add(Dropout(self.dropout))
|
| 50 |
+
|
| 51 |
+
self.generator_model.add(Dense(int(self.number_of_features * 2)))
|
| 52 |
+
self.generator_model.add(LeakyReLU(alpha=self.alpha))
|
| 53 |
+
self.generator_model.add(Dropout(self.dropout))
|
| 54 |
+
|
| 55 |
+
self.generator_model.add(Dense(self.number_of_features, activation='sigmoid'))
|
| 56 |
+
optimizer = Adam(lr=self.learning_rate)
|
| 57 |
+
self.generator_model.compile(loss='categorical_crossentropy', optimizer=optimizer)
|
| 58 |
+
|
| 59 |
+
# self.generator_model.summary()
|
| 60 |
+
|
| 61 |
+
def train_gg(self, x_train, y_train, epochs, batch_size, model_name, data, output_path, to_plot=False):
|
| 62 |
+
"""
|
| 63 |
+
This function running the training stage manually.
|
| 64 |
+
:param output_path: Path to save loss fig
|
| 65 |
+
:param to_plot: Plots the losses if True
|
| 66 |
+
:param x_train: the training set features
|
| 67 |
+
:param y_train: the training set classes
|
| 68 |
+
:param model_name: name of model to save (for generator)
|
| 69 |
+
:param epochs: number of epochs
|
| 70 |
+
:param batch_size: the batch size
|
| 71 |
+
:return: trains the discriminator and generator.
|
| 72 |
+
"""
|
| 73 |
+
|
| 74 |
+
losses_path = os.path.join(self.saved_models_path, f'{model_name}_losses')
|
| 75 |
+
model_file = os.path.join(self.saved_models_path, f'{model_name}_part_2_gen_weights.h5')
|
| 76 |
+
|
| 77 |
+
# First train the discriminator
|
| 78 |
+
self.train_black_box_dis(x_train, y_train)
|
| 79 |
+
self.train_generator(x_train, model_file, epochs, batch_size, losses_path)
|
| 80 |
+
if to_plot:
|
| 81 |
+
self.plot_losses(data, output_path)
|
| 82 |
+
|
| 83 |
+
def train_black_box_dis(self, x_train, y_train):
|
| 84 |
+
"""
|
| 85 |
+
Trains the discriminator and saves it.
|
| 86 |
+
:param x_train: the training set features
|
| 87 |
+
:param y_train: the training set classes
|
| 88 |
+
:return:
|
| 89 |
+
"""
|
| 90 |
+
dis_output = os.path.join(self.saved_models_path, 'black_box_dis_model')
|
| 91 |
+
|
| 92 |
+
if os.path.exists(dis_output):
|
| 93 |
+
# print('Blackbox discriminator already trained')
|
| 94 |
+
with open(dis_output, 'rb') as rf_file:
|
| 95 |
+
self.discriminator_model = pickle.load(rf_file)
|
| 96 |
+
|
| 97 |
+
self.discriminator_model.fit(x_train, y_train)
|
| 98 |
+
with open(dis_output, 'wb') as rf_file:
|
| 99 |
+
pickle.dump(self.discriminator_model, rf_file)
|
| 100 |
+
|
| 101 |
+
def train_generator(self, data, model_path, epochs, start_batch_size, losses_path):
|
| 102 |
+
"""
|
| 103 |
+
Function for training the general generator.
|
| 104 |
+
:param losses_path: The filepath for the loss results
|
| 105 |
+
:param data: The normalized dataset
|
| 106 |
+
:param model_path: The name of the model to save. includes epoch size, batches etc.
|
| 107 |
+
:param epochs: Number of epochs
|
| 108 |
+
:param start_batch_size: Size of batch to use.
|
| 109 |
+
:return: trains the generator, saves it and the losses during training.
|
| 110 |
+
"""
|
| 111 |
+
|
| 112 |
+
if os.path.exists(model_path):
|
| 113 |
+
self.generator_model.load_weights(model_path)
|
| 114 |
+
with open(losses_path, 'rb') as loss_file:
|
| 115 |
+
self.losses = pickle.load(loss_file)
|
| 116 |
+
return
|
| 117 |
+
|
| 118 |
+
for epoch in range(epochs): # iterates over the epochs
|
| 119 |
+
np.random.shuffle(data)
|
| 120 |
+
batch_size = start_batch_size
|
| 121 |
+
for i in tqdm(range(0, data.shape[0], batch_size), ascii=True): # Iterate over batches
|
| 122 |
+
if data.shape[0] - i >= batch_size:
|
| 123 |
+
batch_input = data[i:i + batch_size]
|
| 124 |
+
else: # The last iteration
|
| 125 |
+
batch_input = data[i:]
|
| 126 |
+
batch_size = batch_input.shape[0]
|
| 127 |
+
|
| 128 |
+
g_loss = self.train_generator_on_batch(batch_input)
|
| 129 |
+
self.losses['gen_loss'].append(g_loss)
|
| 130 |
+
|
| 131 |
+
self.save_generator_model(model_path, losses_path)
|
| 132 |
+
|
| 133 |
+
def save_generator_model(self, generator_model_path, losses_path):
|
| 134 |
+
"""
|
| 135 |
+
Saves the model and the loss data with pickle.
|
| 136 |
+
|
| 137 |
+
:param generator_model_path: File path for the generator
|
| 138 |
+
:param losses_path: File path for the losses
|
| 139 |
+
:return:
|
| 140 |
+
"""
|
| 141 |
+
self.generator_model.save_weights(generator_model_path)
|
| 142 |
+
with open(losses_path, 'wb+') as loss_file:
|
| 143 |
+
pickle.dump(self.losses, loss_file)
|
| 144 |
+
|
| 145 |
+
def train_generator_on_batch(self, batch_input):
|
| 146 |
+
"""
|
| 147 |
+
Trains the generator for a single batch. Creates the necessary input, comprised of noise and the real
|
| 148 |
+
probabilities obtained from the black box. Compared to the target output, made of real samples and the
|
| 149 |
+
probabilities made up by the generator.
|
| 150 |
+
:param batch_input:
|
| 151 |
+
:return:
|
| 152 |
+
"""
|
| 153 |
+
batch_size = batch_input.shape[0]
|
| 154 |
+
discriminator_probabilities = self.discriminator_model.predict_proba(batch_input)[:, -1:]
|
| 155 |
+
# noise = randn(self.noise_dim * batch_size).reshape((batch_size, self.noise_dim))
|
| 156 |
+
|
| 157 |
+
noise = randn(batch_size, self.noise_dim)
|
| 158 |
+
gen_model_input = np.hstack([noise, discriminator_probabilities])
|
| 159 |
+
generated_probabilities = self.generator_model.predict(gen_model_input)[:, -1:] # Take only probabilities
|
| 160 |
+
target_output = np.hstack([batch_input, generated_probabilities])
|
| 161 |
+
g_loss = self.generator_model.train_on_batch(gen_model_input, target_output) # The actual training
|
| 162 |
+
|
| 163 |
+
return g_loss
|
| 164 |
+
|
| 165 |
+
def plot_discriminator_results(self, x_test, y_test, data, path):
|
| 166 |
+
"""
|
| 167 |
+
:param x_test: Test set
|
| 168 |
+
:param y_test: Test classes
|
| 169 |
+
:return: Prints the required plots.
|
| 170 |
+
"""
|
| 171 |
+
|
| 172 |
+
blackbox_probs = self.discriminator_model.predict_proba(x_test)
|
| 173 |
+
discriminator_predictions = self.discriminator_model.predict(x_test)
|
| 174 |
+
count_1 = int(np.sum(y_test))
|
| 175 |
+
count_0 = int(y_test.shape[0] - count_1)
|
| 176 |
+
class_data = (['Class 0', 'Class 1'], [count_0, count_1])
|
| 177 |
+
self.plot_data(class_data, path, mode='bar', x_title='Class', title=f'Distribution of classes - {data} dataset')
|
| 178 |
+
self.plot_data(blackbox_probs[:, 0], path, title=f'Probabilities for test set - class 0 - {data} dataset')
|
| 179 |
+
self.plot_data(blackbox_probs[:, 1], path, title=f'Probabilities for test set - class 1 - {data} dataset')
|
| 180 |
+
|
| 181 |
+
min_confidence = blackbox_probs[:, 0].min(), blackbox_probs[:, 1].min()
|
| 182 |
+
max_confidence = blackbox_probs[:, 0].max(), blackbox_probs[:, 1].max()
|
| 183 |
+
mean_confidence = blackbox_probs[:, 0].mean(), blackbox_probs[:, 1].mean()
|
| 184 |
+
|
| 185 |
+
print("Accuracy:", metrics.accuracy_score(y_test, discriminator_predictions))
|
| 186 |
+
for c in [0, 1]:
|
| 187 |
+
print(f'Class {c} - Min confidence: {min_confidence[c]} - Max Confidence: {max_confidence[c]} - '
|
| 188 |
+
f'Mean confidence: {mean_confidence[c]}')
|
| 189 |
+
|
| 190 |
+
def plot_generator_results(self, data, path, num_of_instances=1000):
|
| 191 |
+
"""
|
| 192 |
+
Creates plots for the generator results on 1000 instances.
|
| 193 |
+
:param path:
|
| 194 |
+
:param data: Name of dataset used.
|
| 195 |
+
:param num_of_instances: Number of samples to generate.
|
| 196 |
+
:return:
|
| 197 |
+
"""
|
| 198 |
+
sampled_proba, generated_instances = self.generate_n_samples(num_of_instances)
|
| 199 |
+
|
| 200 |
+
proba_fake = self.discriminator_model.predict_proba(generated_instances[:, :-1])
|
| 201 |
+
for c in [0, 1]:
|
| 202 |
+
title = f'Confidence Score for Class {c} of Fake Samples - {data} dataset'
|
| 203 |
+
self.plot_data(proba_fake[:, c], path, x_title='Confidence Score', title=title)
|
| 204 |
+
|
| 205 |
+
black_box_confidence = proba_fake[:, 1:]
|
| 206 |
+
proba_error = np.abs(sampled_proba - black_box_confidence)
|
| 207 |
+
generated_classes = np.array([int(round(c)) for c in generated_instances[:, -1].tolist()]).reshape(1000, 1)
|
| 208 |
+
proba_stats = np.hstack([sampled_proba, generated_classes, proba_fake[:, :1], proba_fake[:, 1:], proba_error])
|
| 209 |
+
|
| 210 |
+
for c in [0, 1]:
|
| 211 |
+
class_data = proba_stats[proba_stats[:, 1] == c]
|
| 212 |
+
class_data = class_data[class_data[:, 0].argsort()] # Sort it for the plot
|
| 213 |
+
title = f'Error rate for different probabilities, class {c} - {data} dataset'
|
| 214 |
+
self.plot_data((class_data[:, 0], class_data[:, -1]), path, mode='plot', y_title='error rate', title=title)
|
| 215 |
+
|
| 216 |
+
def generate_n_samples(self, n):
|
| 217 |
+
"""
|
| 218 |
+
Functions for generating N samples with a uniformly distribution confidence level.
|
| 219 |
+
:param n: Number of samples
|
| 220 |
+
:return: a tuple of the confidence scores used and the samples created.
|
| 221 |
+
"""
|
| 222 |
+
noise = randn(n, self.noise_dim)
|
| 223 |
+
# confidences = np.sort(np.random.uniform(0, 1, (n, 1)), axis=0)
|
| 224 |
+
confidences = np.random.uniform(0, 1, (n, 1))
|
| 225 |
+
|
| 226 |
+
generator_input = np.hstack([noise, confidences]) # Stick them together
|
| 227 |
+
generated_instances = self.generator_model.predict(generator_input) # Create samples
|
| 228 |
+
|
| 229 |
+
return confidences, generated_instances
|
| 230 |
+
|
| 231 |
+
@staticmethod
|
| 232 |
+
def plot_data(data, path, mode='hist', x_title='Probabilities', y_title='# of Instances', title='Distribution'):
|
| 233 |
+
"""
|
| 234 |
+
:param path: Path to save
|
| 235 |
+
:param mode: Mode to use
|
| 236 |
+
:param y_title: Title of y axis
|
| 237 |
+
:param x_title: Title of x axis
|
| 238 |
+
:param data: Data to plot
|
| 239 |
+
:param title: Title of plot
|
| 240 |
+
:return: Prints a plot
|
| 241 |
+
"""
|
| 242 |
+
plt.clf()
|
| 243 |
+
|
| 244 |
+
if mode == 'hist':
|
| 245 |
+
plt.hist(data)
|
| 246 |
+
elif mode == 'bar':
|
| 247 |
+
plt.bar(data[0], data[1])
|
| 248 |
+
else:
|
| 249 |
+
plt.plot(data[0], data[1])
|
| 250 |
+
|
| 251 |
+
plt.title(title)
|
| 252 |
+
plt.ylabel(y_title)
|
| 253 |
+
plt.xlabel(x_title)
|
| 254 |
+
# plt.show()
|
| 255 |
+
path = os.path.join(path, title)
|
| 256 |
+
plt.savefig(path)
|
| 257 |
+
|
| 258 |
+
def plot_losses(self, data, path):
|
| 259 |
+
"""
|
| 260 |
+
Plot the losses while training
|
| 261 |
+
:return:
|
| 262 |
+
"""
|
| 263 |
+
plt.clf()
|
| 264 |
+
plt.plot(self.losses['gen_loss'])
|
| 265 |
+
plt.title('Model loss')
|
| 266 |
+
plt.ylabel('Loss')
|
| 267 |
+
plt.xlabel('Iteration')
|
| 268 |
+
# plt.show()
|
| 269 |
+
plt.savefig(os.path.join(path, f'{data} dataset - general_generator_loss.png'))
|
| 270 |
+
|
| 271 |
+
def get_error(self, num_of_instances=1000):
|
| 272 |
+
"""
|
| 273 |
+
Calculates the error of the generator we created by measuring the difference between the probability that
|
| 274 |
+
was given as input and the probability of the discriminator on the sample created.
|
| 275 |
+
:param num_of_instances: Number of samples to generate.
|
| 276 |
+
:return: An array of errors.
|
| 277 |
+
"""
|
| 278 |
+
sampled_proba, generated_instances = self.generate_n_samples(num_of_instances)
|
| 279 |
+
proba_fake = self.discriminator_model.predict_proba(generated_instances[:, :-1])
|
| 280 |
+
black_box_confidence = proba_fake[:, 1:]
|
| 281 |
+
return np.abs(sampled_proba - black_box_confidence)
|
| 282 |
+
|
outputs/empty
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
|